In this study:
- International research teams analyzed several hundred thousand samples from 17 whole-exome sequencing studies to identify rare gene variants associated with smoking and alcohol use.
- They identified rare variants in 117 DNA regions as being associated with specific aspects of nicotine and alcohol use, including some in genes already known to influence these traits.
Tobacco and alcohol use are modifiable risk factors that account for more illnesses and deaths in Western societies than any other single risk factor or health condition. Both tobacco and alcohol use are at least in part genetically determined, and elucidating the underlying genetic factors may help prevent or ameliorate the use of these substances and their consequences. Dr. David M. Brazel at the University of Minnesota, Yu Jiang and Jordan Hughey at Penn State College of Medicine, as well as colleagues at more than two dozen institutions in nine countries, recently identified rare gene variants that may influence the risk of tobacco or alcohol use. “Enabled by cost-effective sequencing and genotyping technologies, large-scale genetic datasets with millions of individuals have allowed us to identify numerous novel genes for addiction behavior, the discovery of which has strong public health relevance,” says Dr. Dajiang Liu, one of the study’s senior investigators.
Large Datasets Allow Identification of Rare Gene Variants
Numerous genome-wide association studies (GWAS) have already investigated genes underlying alcohol and tobacco use. However, rare gene variants, especially those that abolish a protein’s normal function, have greater potential to reveal the biological mechanisms underlying such complex traits. They also often have a greater effect on disease risk than common variants. However, few studies have looked at rare variants—commonly defined as occurring in about one out of 1,000 individuals or fewer—because large sample sizes are needed for any reliable conclusions.
The international research team performed the largest meta-analysis to date of rare genetic variants associated with nicotine and alcohol use (see the Figure). They developed novel statistical methods to integrate data from 17 studies for their analyses. Each study had measured rare mutations in participants’ exomes, which are the sections of DNA that encode proteins. The combined sample sizes ranged from 152,348 to 433,216, depending on the specific aspect (i.e., phenotype) of nicotine or alcohol use analyzed. The researchers evaluated five phenotypes: cigarettes smoked per day, pack-years (a measure of total lifetime nicotine exposure), smoking initiation (whether a person had smoked at least 100 cigarettes during their life), age of smoking initiation, and number of alcoholic drinks per week.
DNA Regions Associated With Smoking or Alcohol Use Traits
In total, the team identified rare variants in 171 DNA regions (i.e., loci) throughout the genome that were associated with the five phenotypes. These variants together explained 1.0 percent to 2.2 percent of the phenotypic variance for the five traits and contributed significantly to the heritability of smoking and alcohol use.
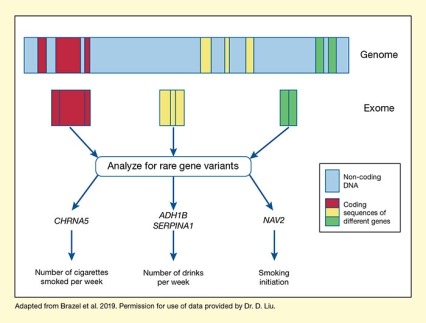
See full text description at end of article.
Some of these variants were in genes previously implicated in smoking and alcohol use. For example, the study detected a strong association between a variant of the gene CHRNA5, which encodes a nicotine receptor, and the number of cigarettes smoked per week. Previous GWAS had also found that a CHRNA5 variant explained about 1 percent of the phenotypic variance of smoking quantity. The new study also suggested an association between a variant in the ADH1B gene, which encodes an alcohol-metabolizing enzyme, and the number of drinks per week. Variants in two other genes, NAV2 and SERPINA1, were significantly associated with smoking initiation and number of drinks per week, respectively (see the Figure).
“From these findings, we hope to figure out mechanistically how these mutations work,” Dr. Liu says. “We also hope to develop medications that can mimic the functions of these genes, and thus help people quit smoking and drinking regardless of whether they carry the mutation or not.”
One limitation of this study, which it shares with almost all meta-analyses of psychiatric traits, was that the vast majority of samples came from individuals with European ancestry; less than 9,000 samples were of African ancestry. This imbalance is significant because genetic risks for many traits vary widely across different populations. Moreover, new medications are usually developed based on the population in which genetic discoveries are made. “Expanding this research into populations of non-European ancestry would be greatly meaningful for understanding how genetics influence smoking and alcohol addiction in worldwide populations,” Dr. Liu says.
This study was supported by NIDA grants DA037904, DA040177, and DA017637-13.
- Text Description of Figure
-
The figure illustrates the design and main findings of the study to identify rare gene variants related to alcohol and smoking phenotypes. A broad bar at the top represents the genomic DNA. Light blue areas indicate non-coding DNA regions, which are interspersed with coding DNA sequences of three hypothetical genes, represented by three red sections on the left, three yellow sections in the middle, and two green sections on the right. The three red, yellow, and green boxes below this bar represent the combined coding sequences for the three hypothetical genes as they are found in the exome. Three blue arrows pointing from the three colored boxes to the text box indicate that the researchers analyzed these exome sequences for rare genetic variants. As indicated by three blue arrows pointing away from the textbox, these analyses identified variants in four genes that were associated with specific phenotypes (indicated by blue arrows pointing down from the gene names to the functions). These included CHRNA5, which was associated with number of cigarettes smoked per week, on the left; ADH1B and SERPINA1, which both were associated with number of drinks per week, in the middle; and NAV2, which was associated with smoking initiation, on the right.
Source:
- Brazel, D.M., Jiang, Y., Hughey, J.M., et al. Exome Chip Meta-analysis Fine Maps Causal Variants and Elucidates the Genetic Architecture of Rare Coding Variants in Smoking and Alcohol Use. Biological Psychiatry. 85(11):946-955, 2019.